Effective Strategies for Successful Hit Identification in Drug Discovery
What’s the optimal hit identification approach to enable success?
Here, we take a look at knowledge-based hit generation, HTS, big data virtual screening and fragment-based screening options.

Knowledge-Based Hit Generation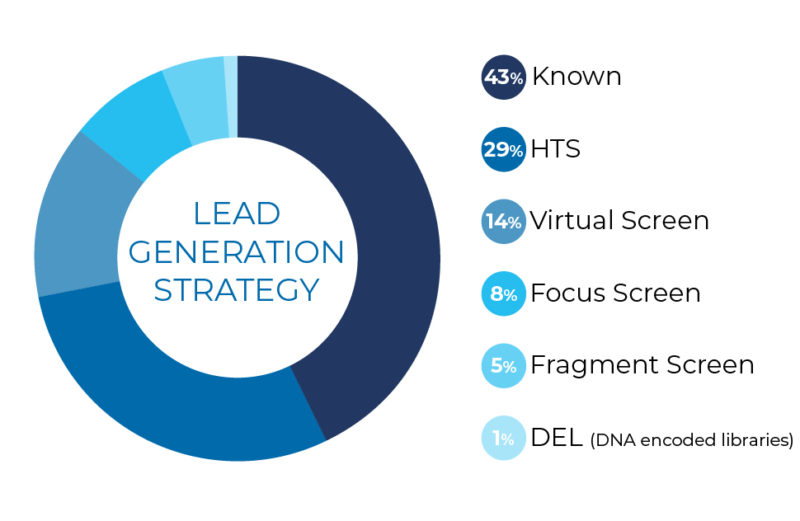
According to a literature survey [1], around 40% of small molecule drug candidates come from chemical matter generated by a knowledge-based approach.
Many of the drug candidates Sygnature Discovery has delivered to our clients started this way using extensive database searching, pharmacophore matching and scaffold hopping computational approaches. This approach can reduce the time taken to get to candidate selection, but does carry a greater risk of others finding the same IP space.
High Throughput Screening (HTS)
The next most common origin of drug candidates is High Throughput Screening (HTS), using automated screening of plate-based, diverse compound libraries.
In the past this technique often relied on poorly curated and out-of-date compound decks derived from historical collections acquired from a company’s medicinal chemistry output, augmented by commercial compounds or combi-chem libraries.
Sygnature’s lead-like screening library (LeadFinder), selected from recently synthesized compounds from multiple commercial vendors, stands out as a fresh opportunity to mine for better and more progressable hits. The in silico selection process based on scaffold diversity and physico-chemical properties, has been further refined by our medicinal chemists to ensure the compounds represent viable starting matter for a project.
Our library, together with highly integrated compound management and data processing systems and state-of-the-art automation, provides high-fidelity cellular and biochemical HTS screens that have already delivered multiple hit series for client projects.
Large Database Virtual Screening
Recent advances in 3D-searching technology (Open Eye’s Fast-ROCS) enable us to search very large compound databases, such as the Enamine REAL database of 0.75 billion synthesizable drug-like molecules, in a remarkably short time.
Using this technology on commercially available compounds or compounds that are synthetically enabled is a very effective way of enriching an HTS with compounds based on structure, ligands or fragments. It can also be used to select smaller focused compound sets for lower throughput high-content or phenotypic screening.
Fragment-Based Screening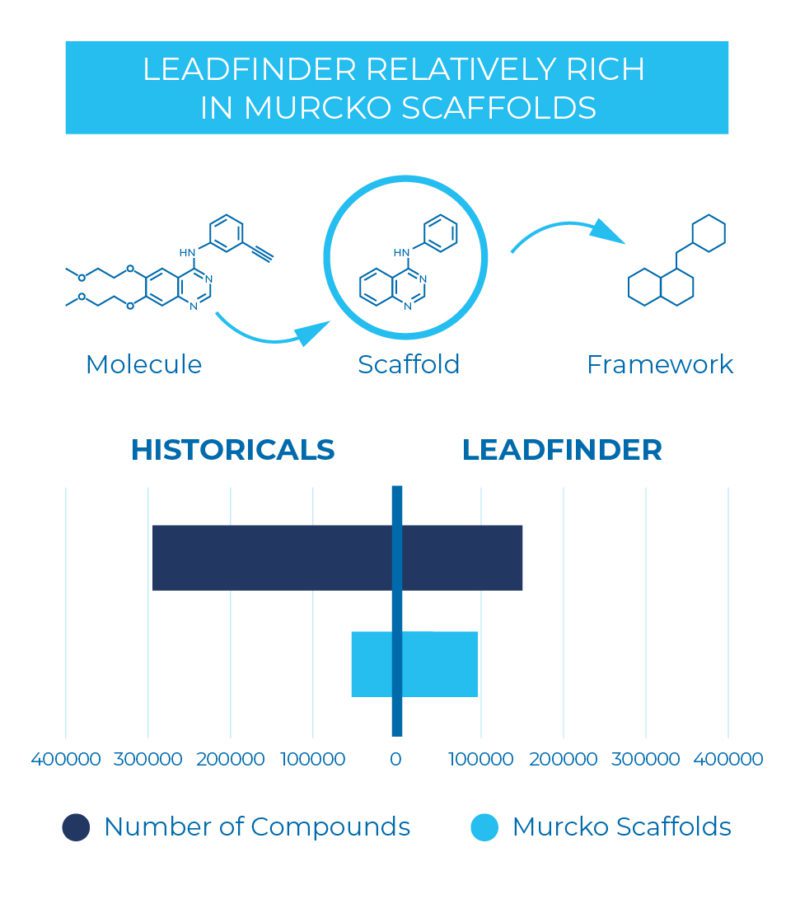
Finally, where structure of the protein target or a close homologue is known, a structure-based approach using flexible docking of commercial or synthetically enabled compounds can be effective.
Screening of a well-designed fragment library, like Sygnature’s 1,700 strong proprietary collection, can be used to assess drugability [2], provide small starting fragment hits or feed into virtual screening efforts in selection of larger molecules likely to bind the target protein.
Such highly focused structure-based screens can take advantage of lower throughput direct binding measurements using a variety of biophysical methods such as SPR and MST. Confirmation of hits using orthogonal methods such as NMR or X-ray crystallography is then used to give information about the likely binding mode and to facilitate guided fragment extension in a highly efficient manner.
All of the above described methods for hit discovery are in use at Sygnature Discovery and have each or in combination delivered exciting hit series for client projects.
If you would like to know what methods might be advantageous in your project, our expert scientists will be happy to discuss the merits of each approach, so please feel free to get in touch via our simple contact form.
1. Dean G. Brown, Jonas Boström; J. Med. Chem. 2018, 61, 21, 9442-9468
2. Molly Chilton, Ben Clennell, Fredrik Edfeldt, Stefan Geschwindner; J. Med. Chem. 2017, 60, 12, 4923-4931
