Sygnature Discovery announces collaboration with software firm SilcsBio
Sygnature Discovery announces today a collaboration with SilcsBio, a scientific software company focused on high-throughput fully atomistic simulations of protein targets and protein-ligand structures.
SilcsBio has provided Sygnature with access to its SILCS (Site Identification by Ligand Competitive Saturation) and SSFEP (Single Step Free Energy Perturbation) software. Sygnature’s researchers are now able to offer clients the benefits of these powerful techniques in their drug discovery projects.
The SILCS software identifies binding site hot spots on a protein’s surface. This could, potentially, include ‘cryptic pockets’ – binding sites for which little structural information is available. SILCS methodology involves performing molecular dynamics simulations on a protein in an aqueous solution of chemically diverse small molecules. This generates probability distributions of fragment types, termed FragMaps. These FragMaps can be used rapidly and with minimal computational expense to predict or refine ligand binding poses, quantitatively score ligands in the binding pocket, and generate multiple receptor-based pharmacophore models for use in virtual screening campaigns. This has been shown to be a more effective technique than standard docking studies or receptor-based pharmacophore modelling.
In a complementary manner, the SSFEP software has demonstrated success in ranking compound binding affinities to target proteins. In particular, the SSFEP approach is about 1000-fold faster than the more widely used FEP methodology.
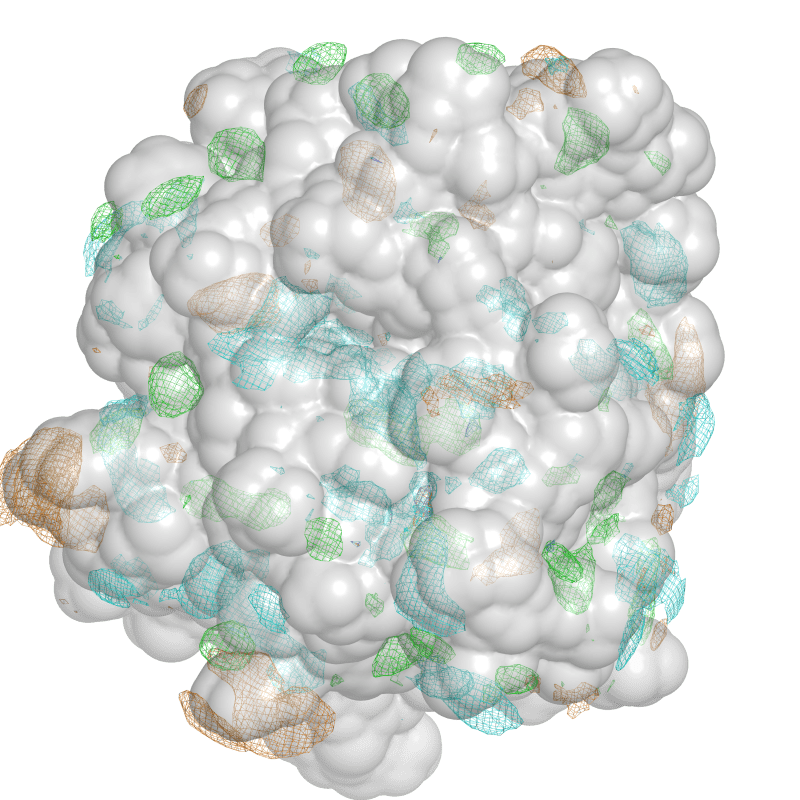
Figure of FragMaps for a given protein structure
It achieves this speed by post-processing molecular dynamics simulation data of a ligand, and using it to estimate the alchemical free energy change of chemically modifying that ligand.
Such a rapid assessment of the drug design team’s ideas could be advantageous in speeding up the design-test-optimise cycle. Being able to evaluate and prioritise design ideas in just a few days can have a significant impact in structure-based design projects.
In the past six months, Sygnature has applied the SilcsBio software suite successfully in various hit-to-lead and lead optimisation projects. The company believes these new software tools will offer substantial benefits to its customers, not least because of the time savings the software can provide.
EXAMPLES OF PREVIOUS SILCSBIO’S SOFTWARE SUCCESSES INCLUDE:
- Cryptic/transient binding pocket detection in GPCRs, Nuclear receptors & other transmembrane proteins
- Revealed previously unknown binding pockets, allowing customers to design of more potent ligands
- Predicted crystallographic binding modes of customers’ newly designed inhibitors
- Provided crucial insights to drive medicinal chemistry refinement of BCL6 inhibitors
- 2019 NIH award to fund expansion of SILCS software to enable its use with biological therapeutics, especially for excipient design
“We’re delighted to be collaborating with Sygnature Discovery to bring the advantages of our drug design software to Sygnature’s projects and customers. This collaboration is very meaningful to us as Sygnature continues to be a great partner and it opens the door to new markets for SilcsBio”, says Ken Malone, Chief Executive Officer of SilcsBio.
Bill Tatsis, Computational Chemist at Sygnature Discovery commented “SilcsBio software suite, with its robust physics-based algorithms and user-friendly interface, provides great leverage in the structure-based drug design area and enables us to rapidly validate new ideas/hypothesis”.
If you have any questions, would like to discuss our capabilities or drug discovery in general then we’d love to hear from you. You can get in touch by using any of the contact forms or by emailing us at info@sygnaturediscovery.com
