Fragment-based Lead Discovery to Identify Novel Bromodomain-Containing Protein 3 Binders
Abstract
Sygnature Discovery has designed and synthesised a proprietary fragment library with a high degree of novelty that covers a wide range of chemical space. Comprised of over 900 compounds, most of which are not commercially available (>60%), the library was designed using the ‘rule of 3’ as a guideline and with a strong emphasis on diversity.
To demonstrate its effectiveness in fragment-based drug discovery, the library was screened against an epigenetic target with little previous literature on drug discovery, Bromodomain-containing protein 3 (BRD3, also known as RING3L). Like other members of the Bromodomain and Extra-Terminalmotif (BET) family, BRD3 contains two tandem homologous bromodomains and an extra terminal motif. It functions by binding acetylated lysine residues on chromatin and transcriptional regulators, exemplified by the role it plays in the regulation of transcription by promoting the binding of GATA. Due to this involvement in regulation, BET members often play a role in several types of cancer. BRD3, in particular, is associated with a number of disease phenotypes. For example, depletion of BRD3 slows growth in cancer models including prostate cancer and medulloblastoma and BRD3 has been implicated in NUT midline carcinoma (NMT).
While there are a number of available pan-BET inhibitors, the design of specific BRD3 inhibitors may lead to a beneficial clinical outcome with reduced off-target effects. However, bromodomains of the BET family have a high degree of structural similarity, especially in the acetylated lysine binding pocket, making the design of selective inhibitors problematic. BRD3, therefore, represents an intriguing and challenging drug discovery target.
Introduction
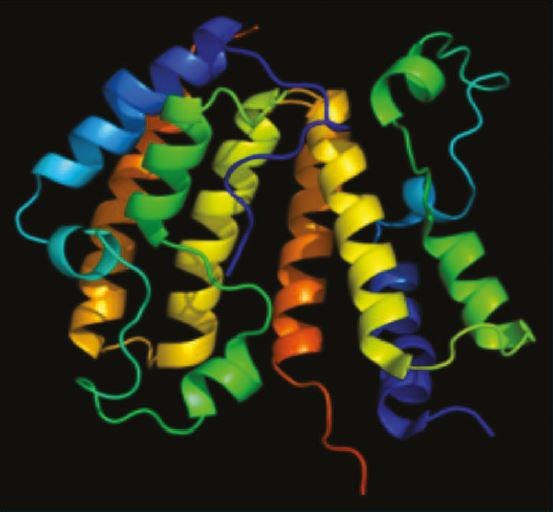
Figure 1: PDB rendering based on 2nxb
Fragment-based approaches have been applied successfully to the discovery and optimization of ligands for a wide range of targets. Fragment-based lead discovery (FBLD) is proving particularly valuable for bromodomain (BRD)-containing proteins, an emerging family of epigenetic regulators with potential as targets for cancer, inflammation and multiple sclerosis [1]. These small proteins (~110 amino acids) are able to recognize and read the acetylated lysine (KAc) modifications on histones and are subsequently responsible for transducing and translating signals into various normal or abnormal phenotypes.
The strongest phenotypic changes to date have been found with inhibitors of the BET (Bromodomain and Extra Terminal) family, which includes BRD2 (also known as Fsrg1 or RING3), BRD3 (Fsrg2, ORFX), BRD4 (Fsrg4, MCAP/HUNK1), and BRDT (Fsrg3, BRD6). The vast majority of FBLD/bromodomain projects to date have involved identifying fragments binding to BRD4 [2] or BRD2 [3] and have resulted in a number of potent pan-BET inhibitors. However, there is growing evidence that compounds able to inhibit just BRD3 or BRD2/BRD3, as opposed to BRD4 or pan-BET inhibitors, will result in different phenotypic outcomes. For example, the absence of BRD3 results in reduced growth of prostate cancer ,whereas the chromosomal translocation of BRD3with the NUT gene is associated with NUT midline carcinoma [6].
By exploiting our in-house fragment library, various biophysical techniques including surface plasmon resonance (SPR), and protein X-ray crystallography, we have identified and characterized fragments that bind at the acetyl-lysine binding site of the first bromodomain in BRD3 (BRD3/BD1). Further chemical elaboration of these fragments could lead to a BRD3 or BRD2/3 specific molecule that could be utilized as a probe for elucidating the individual roles of the members of the BET-family.
Fragment Library Design
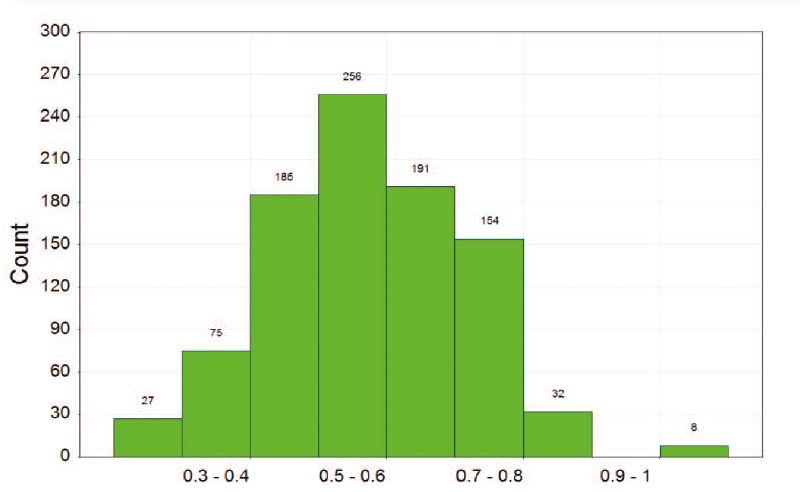
Figure 2: Max Tanimoto coefficient (mean: 0.57)
Sygnature Discovery has designed and synthesised a diverse fragment library, readily amenable to medicinal chemists, with each fragment containing at least one point of diversity for elaboration. To aid identification of true hits and suggest early SAR the library contains a small number of analogues (1-4) for each fragment.
The library has been clean screened to remove any PAINS (pan-assay interference compounds) or reactives and all fragments have been purity checked (> 95%purity) using NMR and LCMS.
The library was designed using the ‘rule of 3’ as guidelines rather than applying strict cut-offs. Therefore, while the average properties fall within the ‘rules’, the chemical space is extended.
Library Characterisation
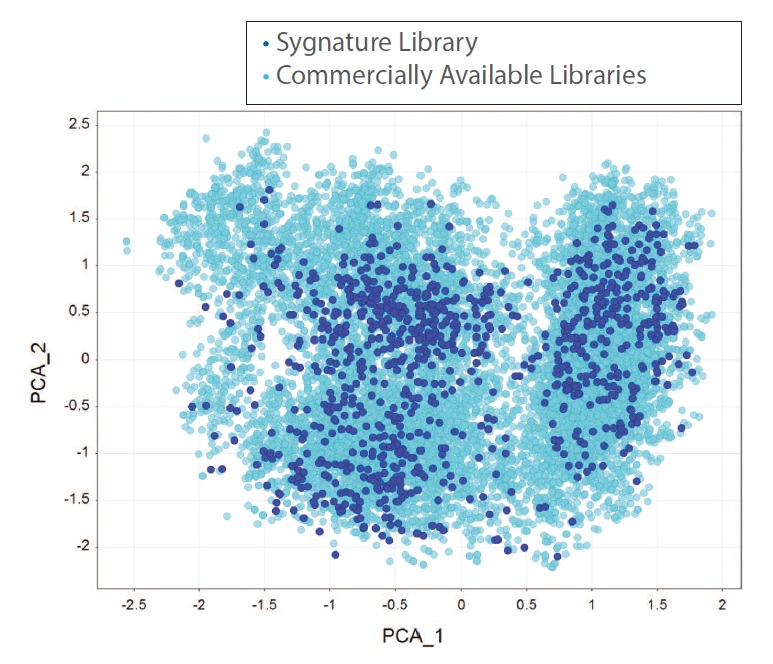
Figure 3: PCA comparison of Sygnature fragment library with 10,650 fragments from commercially available fragment libraries with measured solubility >1 mM
Internal diversity of the library and coverage of chemical space were assessed using circular fingerprints (FeatMorgan, radius 3).
The distribution of the closest Tanimoto similarity for each compound highlights that Sygnature’s Fragment Library contains both highly diverse compounds and clusters of analogues.
With ~900 fragments, the Sygnature Fragment Library provides very good coverage of the soluble fragment chemical space. There is very little structural overlap between Sygnature’s fragment library and commercially available fragment libraries as most of the fragments (> 60%)were designed and synthesised in-house.
Protein Production
In collaboration with Peak Proteins, recombinant BRD3/BD1, containing an N-terminal 6xHis-tag, was cloned and expressed in E.coli. Initial SPR assays using the pan-BET inhibitor I-BET 151 demonstrated the protein was fully functional with the expected compound binding behaviour, and was therefore suitable for screening.
Screening
Surface Plasmon Resonance (SPR)
908 fragments were screened at 250 μM, 1% DMSO against BRD3/BD1, BRD4/BD1 (for selectivity/confirmation) and human Carbonic anhydrase II (for specificity).
Fragment actives were initially identified by having a normalised response of 2x standard deviation of blank injections. This resulted in 275 actives that were further interrogated for sensorgram shape (i.e. square wave), effects on baseline and potential for superstoichiometric binding. Using these criteria we further reduced the actives to 148. By performing concentration response curves to calculate KD for each of these, it was found that 20 fragments had repeatable, saturating dose responses. Nine fragments were chosen for X-ray structural analysis, based on the reproducibility of the dose responses with solid stock.
X-Ray Crystallography
X-ray crystallography was performed by Peak Proteins. BRD3/BD1 apo-crystals were soaked with single fragments (10-20 mM) and diffraction data collected at the Diamond Light Source (Oxford, UK). Of the nine crystals analysed, six were found to contain fragments with unambiguous binding modes, three of which are highlighted below.
Two fragment analogues SYG-00003236 and 3237 gave comparable KD by SPR and have been shown to bind BRD3D1 in a similar manner (Figure 5A and 5B). SYG-00000127 had the highest affinity with a KD < 10 μM. The increased potency is likely to be derived from five H-bonds to water molecules (Figure 5C).
Conclusion
- A fragment screening programme to identify BRD3/BD1 binders was successfully completed using our proprietary fragment library, SPR screening and X-ray crystallography
- Of the nine fragments identified as binders from the SPR screen, six gave liganded structures when soaked with BRD3/BD1 construct
- The highest affinity fragment had a KD of < 10 μM with five fragment-water H-bonds
- Next steps could include fragment linking, hit expansion, SAR refinement and characterisation of selective compounds in biological assays to eventually develop tool compounds to interrogate the role of BRD3 in disease phenotypes.

